Use Parsl to create concurrent computational chemistry workflows
A few weeks ago I posted my first blog post where I explained how to use the Quantum Accelerator (Quacc) package to run DFT calculations with Quantum Espresso. Quacc allows you to link your favorite DFT code to a workflow manager, in this blog post I will show you how to run Quantum Espresso calculations using Quacc and the Parsl workflow engine to create computational chemistry workflows. The main point being to run multiple calculations concurrently on your local machine or on a high-performance computing cluster.
Quacc & Parsl tutorial

If you never run a Quacc calculation before, I recommend you to either read the Quacc documentation or read my previous blog post. For now, I will assume you have Quacc installed and that you have set up the configuration file correctly.
Before we start, you will need to install Parsl:
pip install parsl
Now that you have Parsl installed, you can configure Quacc to use this workflow engine by changing your .quacc.yaml
quacc set WORKFLOW_ENGINE parsl
The aim of the tutorial is to compute the phonon dispersion of multiple bulk crystals using the espresso ph.x code. The grid_phonon_flow available in Quacc will make this problem embarrassingly parallel by performing each representation of each q-points separately. Using Parsl these calculations are then done concurrently automatically depending on the resources available.
The base code presented in this tutorial is fairly simple, the most complex part being the configuration. It first runs a variable-cell relaxation, and then uses the results to run the phonon calculation. From there it extracts the force constant using q2r.x and then call matdyn.x to compute the phonon dispersion on an arbitrary q-point grid. For now, let’s skip the technical details and focus on the functions being called:
@subflow
def grid_phonon_dos_subflow(atoms_list):
results = []
for atoms in atoms_list:
grid_phonon_results = grid_phonon_flow(
atoms,
job_params=grid_phonon_params,
)
q2r_results = q2r_job(grid_phonon_results["dir_name"], **q2r_params)
matdyn_results = matdyn_job(q2r_results["dir_name"], **matdyn_params)
results.append(matdyn_results)
return results
-
grid_phonon_flowis a pre-made@flowin Quacc, that contains multiple@job, details can be found in the documentation. -
q2r_jobis a pre-made@jobthat runs theq2r.xbinary. -
matdyn_jobis a pre-made@jobthat runs thematdyn.xbinary.
The aim is then simple, we take a list of ASE Atoms objects, and we run this custom workflow for each of them. Each results are then stored in a list and returned. Now that we have to core of the workflow we have to configure Parsl.
Setting up Parsl
Parsl runs locally and will submit each job to an HPC scheduler (Slurm in this tutorial). All the options need to be specified via the Parsl configuration.
from parsl.config import Config
from parsl.executors import HighThroughputExecutor
from parsl.launchers import SimpleLauncher
from parsl.providers import SlurmProvider
CONFIG = Config(
executors=[
HighThroughputExecutor(
label="short-large",
max_workers=32, # max number of @job to run concurently.
cores_per_worker=1.0e-6, # number of cores per worker, < 1 to oversubscribe.
provider=SlurmProvider(
account="e89-soto", # your account code.
qos="short", # which quality of service.
worker_init=worker_init, # bash lines that will run before each @job
walltime="00:20:00",
nodes_per_block=32, # nodes for each slurm job (block)
cores_per_node=1, # How many threads to use per parsl worker.
partition="standard",
init_blocks=0, # To be kept to 0, especially if you are using different executors.
max_blocks=1, # Max number of slurm_jobs.
launcher=SimpleLauncher(), # Control where Parsl will live,
),
),
]
)
parsl.load(CONFIG)
The terminology is a bit different from what you might be used to, here is a quick summary:
-
blocksis the number of actual Slurm jobs. You have total control over that by using the keywordsinit_blocks,min_blocksandmax_blocks. -
workersis also a concept introduced by Parsl, this is a computing unit that will take care of running a@job. In practice, this number will dictate how many of your@job’s can run concurrently. -
launcherdictates how the Parsl command is wrapped before being launched, theSimpleLauncherused here means that it does not use any wrapping; the Parsl process will only run on the mother node. TheSingleNodeLauncherwill run the parsl process on each node, and might be used as well. -
cores_per_workeris the number of cores that will be used by each worker. In the case of computational chemistry, you will often see that it is set to something very low, this means that multiple workers can live on the same threads. There is no need to tweakcores_per_node.
These parallelization options are not to be seen as the ones that will affect your individual DFT calculations but more as a total parallelization that will dictate how many resources this executor can use. Your DFT calculations are then dispatched inside this executor, their parallelization will be specified later.
worker_init should have everything you need to run espresso, the machine I use does not have access to the home system from the compute nodes, so I need to to load a ‘scratch’ bashrc that contains the conda init. This is done by adding the following lines:
worker_init = """
source /path/inside/scratch/.bashrc # Load your bashrc that contains the conda init.
conda activate quacc # Activate the conda environment.
export QUACC_CONFIG_FILE=/path/inside/scratch/.quacc.yaml # Quacc config in scratch space.
module load PrgEnv-gnu
module load cray-fftw cray-hdf5-parallel
export LD_LIBRARY_PATH=$CRAY_LD_LIBRARY_PATH:$LD_LIBRARY_PATH
"""
Specifying parameters and parallelization
Before running the workflow you need to setup the parameters that will be sent to each flow and job, namely the grid_phonon_params, matdyn_params, and q2r_params. For the parallelization, currently, the Quacc Espresso interface uses the new parallel_info mechanism introduced in ASE. Here we will use the srun command with its options. This is done by setting the parallel_info variable in the grid_phonon_params and matdyn_params dictionaries.
parallel_info = {
"binary": "srun",
"-vv": True,
"--hint=nomultithread": True,
"--distribution=block:block": True,
"-N": 1,
"-n": 128,
"-c": 1,
}
# pw.x parameters
pw_input_data = {
"control": {"forc_conv_thr": 0.0001},
"system": {
"occupations": "smearing",
"degauss": 0.01,
"smearing": "cold",
},
"electrons": {"conv_thr": 1e-12, "mixing_mode": "TF", "mixing_beta": 0.7},
"ions": {},
"cell": {"press_conv_thr": 0.1},
}
# ph.x parameters
ph_input_data = {
"inputph": {
"tr2_ph": 1.0e-12,
"alpha_mix(1)": 0.1,
"nmix_ph": 8,
"ldisp": True,
"nq1": 5,
"nq2": 5,
"nq3": 5,
},
}
# matdyn.x parameters
matdyn_input_data = {
"input": {
"asr": "crystal",
"dos": True,
"nk1": 32,
"nk2": 32,
"nk3": 32,
"deltaE": 0.5,
},
}
# let's gather all the parameters to send to grid_phonon_flow
grid_phonon_params = {
"relax_job": {
"input_data": pw_input_data,
"kspacing": 0.1,
"relax_cell": True,
"parallel_info": parallel_info,
},
"ph_job": {
"input_data": ph_input_data,
"parallel_info": parallel_info,
},
}
# let's gather all the parameters to send to q2r_job and matdyn_job
matdyn_params = {
"input_data": matdyn_input_data,
"parallel_info": parallel_info,
}
q2r_params = {
"parallel_info": parallel_info,
}
All these lines and config are to be placed before the @subflow “grid_phonon_dos_subflow”. You can access the complete script on my GitHub page. At this point, you can create an atoms list of bulk crystals and call the function.
from ase.build import bulk
atoms_list = [
bulk("Al", cubic=True),
bulk("Cu", cubic=True),
bulk("Ag", cubic=True),
bulk("Au", cubic=True),
bulk("Ni", cubic=True),
bulk("Pd", cubic=True),
bulk("Pt", cubic=True),
bulk("Li", cubic=True),
]
future = grid_phonon_dos_subflow(atoms_list)
At this stage, running the Python script doesn’t produce any output. This is because Parsl is now handling the function call, and it is only generating the Directed Acyclic Graph (DAG), which is an internal representation of the workflow. The returned object is a future object. To actually execute the workflow, the future object needs to be resolved by calling the Future.result() method.
results = future.result()
If you have important disparities between the size of your jobs, Parsl allows you to define multiple executors. To do this, you just need to add a new executor to the CONFIG object. I will not show it here, but an example is available on my GitHub. Using Quacc each job can be linked to a single executor to make sure to run your jobs using the resources you want.
q2r_job_custom = redecorate(q2r_job, job(executors=["q2r"]))
matdyn_job_custom = redecorate(matdyn_job, job(executors=["matdyn"]))
What will happen exactly?
Parsl will construct the workflow internally and be able to manage the intrinsic dependencies between the jobs. As a consequence, two jobs that do not depend on each other can possibly run at the same time. To make things clearer I made the diagram below that summarizes what is happening.
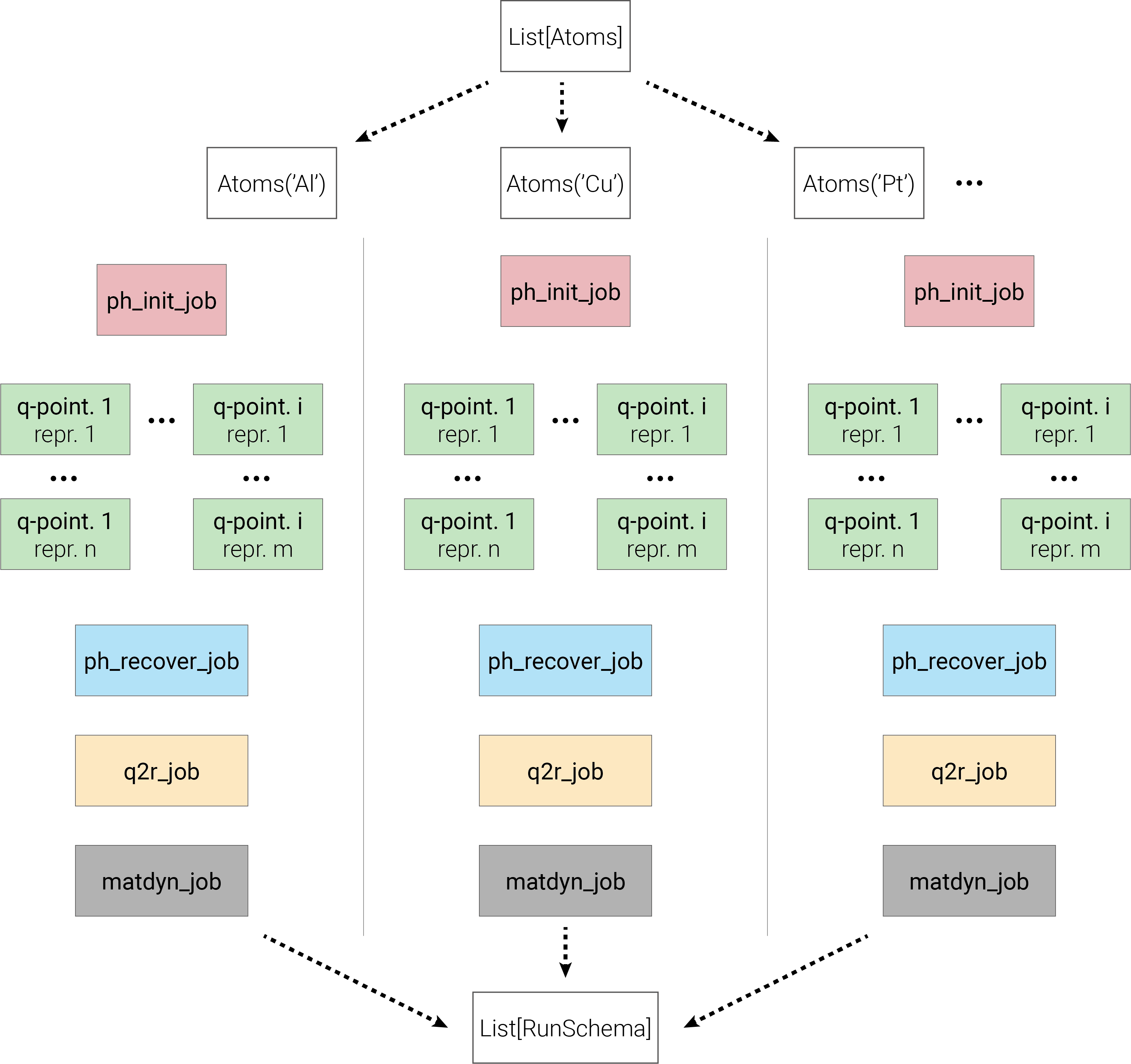
Depending on the number of workers you have set, you will see as many concurrent calculations, other tasks will be queued and run as soon as a worker is available.
Where is the output?
Using Parsl Quacc’s output is still located depending on the settings in the .quacc.yaml file, as described in the documentation. Nothing changed there.
Parsl will also output useful information, by default this is located in a directory named runinfo in the current working directory. If you find yourself struggling to debug your code when using Parsl, check out their documentation.
What if one job fails?
Parsl is able to handle this, if a job fails, every other jobs that do not have the failed job as a dependency will run. This is a very powerful feature that allows to run high-throughput workflows without having to worry too much about the stability of the system. Be aware that the future.result() will still raise an exception if a job fails.
That’s it for this blog post! You will probably need to tweak the configuration to fit your needs, but this should give you a good starting point. Do not hesitate to contact the Parsl support on Slack if you need help, they are very responsive, otherwise, feel free to ask them in the comments below.